Agrigenomics Information Database
Crops such as Rice Genome Database
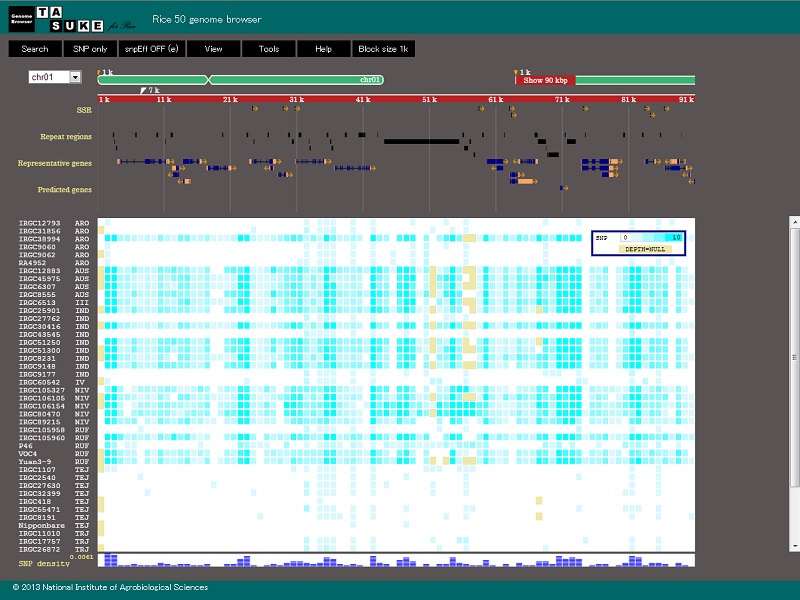 | Rice 50 genome browser |
| Taxonomy Name: Oryza sativa | |
| Description: This is the genome browser 'TASUKE'. This browser shows genome wide variant and coverage depth of re-sequencing data as well as annotation information such as genes, repeats, markers and the rest. Number of variants and depth of coverage are shown by gradational colors in a block whose size is variable from 1bp to 100kb. |
 | MEGANTE |
| Taxonomy Name: | |
| Description: MEGANTE is a web service that makes plant genome annotation easy for non-bioinformaticians. The annotation is visualized with a genome browser and the results can be downloaded in Microsoft Excel format. |
 | Komugi GSP |
| Taxonomy Name: Triticum aestivum | |
| Description: On this web site you can find novels, such as published manuscripts, presentations in conferences/meetings, of the activities for the Komugi Genome Sequence Program, which is a genome analysis program of wheat in Japan. And also you can access to our data including the genome sequences, transcriptomes, annotation data. |
 | bex-db |
| Taxonomy Name: barley (Hordeum vulgare) | |
| Description: Database for barley Full-Length cDNAs and expression data |
 | DAIZUbase |
| Taxonomy Name: Glycine max | |
| Description: DAIZUbase is an integrated soybean genome database and data mining tool, consists of 2 map browsers, a gene viewer, and BLAST search system. |
 | RiceXPro |
| Taxonomy Name: Oryza sativa | |
| Description: The Rice Expression Profile Database (RiceXPro) is a repository of gene expression profiles derived from microarray analysis of tissues/organs encompassing the entire growth of the rice plant under natural field conditions, rice seedlings treated with various phytohormones, and specific cell types/tissues isolated by laser microdissection (LMD). |
 | RiceFREND |
| Taxonomy Name: Oryza sativa | |
| Description: RiceFREND is a gene coexpression database in rice based on a large collection of microarray data derived from various tissues / organs at different stages of growth and development under natural field conditions, and rice plants treated with various phytohormones. The gene expression data used for coexpression analysis can be accessed in the RiceXPro, a repository of gene expression profiles obtained by microarray analysis. RiceFREND is aimed at providing a platform for identification of functionally related genes in various biological pathways and/or metabolic processes. |
 | SALAD Database |
| Taxonomy Name: Oryza sativa | |
| Description: A motif-based database of protein annotations for plant comparative genomics. |
 | FiT-DB |
| Taxonomy Name: Oryza sativa | |
| Description: FiT-DB(Field Transcriptome Database) provides results of statistical modeling of transcriptomic dynamics in the field condition. You can find expression data and parameters of the best-fit model for each gene of Oryza sativa. |
 | RAP-DB |
| Taxonomy Name: Oryza sativa | |
| Description: The Rice Annotation Project (RAP) was conceptualized in 2004 upon the completion of the Oryza sativa ssp. japonica cv. Nipponbare genome sequencing by the International Rice Genome Sequencing Project with the aim of providing the scientific community with an accurate and timely annotation of the rice genome sequence. One of the major objectives of this project is to facilitate a comprehensive analysis of the genome structure and function of rice on the basis of the annotation. |
 | Mutant Panel |
| Taxonomy Name: Oryza sativa Japonica Group | |
| Description: This page is an in silico rice mutant screening page by BLAST search against flanking sequences* from our mutant lines induced by rice retrotransposon Tos17*. You are also able to browse through information about not only insertion points on the rice genome but also phenotype of each mutant line. You can request seeds of the mutant line identified by the screening up to 20 seeds/line. |
 | Q-TARO |
| Taxonomy Name: Oryza sativa | |
| Description: This is a database of Rice QTL information extracted from published research papers. From 1214 reports, we selected 5096 QTLs. The positions of these QTLs were estimated from the physical positions of either two flanking markers (the genomic positions of the distal ends of the two markers) or a co-segregated single marker (between the start and end positions on the genome). If the QTLs were isolated by map-based cloning, the position of the BAC/PAC clone or gene locus was used. By screening for redundancy of traits at the same physical positions, as of March 31 of 2008, we selected 1051 QTLs extracted from 463 reports as representative QTLs. To arrange QTL information, we constructed QTL database (QTL Annotation Rice Online database; Q-TARO, http://qtaro.abr.affrc.go.jp/) consists of two web interfaces. One interface is a table containing information on the mapping of each QTL and its genetic parameters. The other interface is a genome viewer for viewing genomic locations of the QTLs. Currently, update of QTL information is not continued. |
 | OGRO |
| Taxonomy Name: Oryza sativa | |
| Description: Information on the functionally characterized genes compiled is now available in the Overview of Functionally Characterized Genes in Rice Online database (OGRO) on the Q-TARO website (http://qtaro.abr.affrc.go.jp/ogro webcite). The database has two interfaces: a table containing gene information, and a genome viewer that allows users to compare the locations of QTLs and functionally characterized genes. OGRO on Q-TARO will facilitate a candidate-gene approach to identifying the genes responsible for QTLs. Because the QTL descriptions in Q-TARO contain information on agronomic traits, such comparisons will also facilitate the annotation of functionally characterized genes in terms of their effects on traits important for rice breeding. The increasing amount of information on rice gene function being generated from mutant panels and other types of studies will make the OGRO database even more valuable in the future. |
 | ineweb |
| Taxonomy Name: Oryza sativa Japonica Group | |
| Description: The database was recorded and characteristics of rice breed information. |
 | PLACE |
| Taxonomy Name: Embryophyta | |
| Description: PLACE is a database of motifs found in plant cis-acting regulatory DNA elements based on previously published reports on vascular plants including the variations that have been identified in these motifs in other genes or in other plant species in later publications. The database also contains brief descriptions of each motif, literatures with PubMed links, and the corresponding accession numbers in the DDBJ/EMBL/GenBank. |
 | SGD |
| Taxonomy Name: Cryptomeria japonica, Chamaecyparis obtusa | |
| Description: We have conducted genome project of sugi (Cryptomeria japonica) since 1998 and could obtain EST information, DNA markers, linkage maps, and genetic diversity in this species. We have also studied another Japanese conifer, Chamaecyparis obtusa, which is closely related to C. japonica. Therefore, we provide these genome information in this site. |
 | GFSelector |
| Taxonomy Name: Oryza sativa | |
| Description: Gene Function Selector (GFSelector) is a program to predict gene using multiple analysis tools from genome sequences. This program is used in the Rice Genome Automated Annotation System (RiceGAAS). This web site contains the outline explanation of GFSelector and links to an analysis result of the rice genome sequence. |
 | RGP |
| Taxonomy Name: Oryza sativa | |
| Description: High-density molecular genetic map of the rice genome, marshalling map yeast artificial chromosome (YAC), EST sequence information of 65000 pieces, and gene expression maps positioned to YAC marshal on a map EST these has been published. |
 | RGP: RICE GENETIC MAP IN NATURE GENETICS |
| Taxonomy Name: Oryza sativa | |
| Description: This database contains the 1993 version of 1383 RFLP marker information which were used for rice linkage map construction. |
 | RGP gmap98 |
| Taxonomy Name: Oryza sativa | |
| Description: This database contains the 1998 version of 2275 RFLP marker information which were used for rice linkage map construction. |
 | RGP gmap2000 |
| Taxonomy Name: Oryza sativa | |
| Description: This database contains the 2000 version of 3267 RFLP marker information which were used for rice linkage map construction. |
 | RGP caps |
| Taxonomy Name: Oryza sativa | |
| Description: This database contains 332 STS and CAPS marker information which were used for rice linkage map construction. |
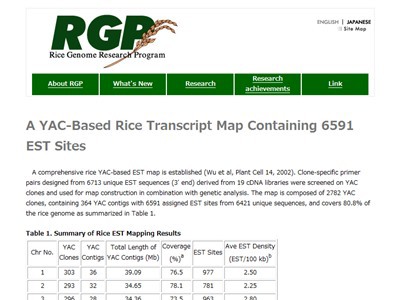 | RGP estmap2001 |
| Taxonomy Name: Oryza sativa | |
| Description: This database contains rice gene expression map which mapped 6591 expressed sequence tag (EST) on yeast artificial chromosome (YAC) alignment map. |
 | RGP physicalmap |
| Taxonomy Name: Oryza sativa | |
| Description: This database contains the yeast artificial chromosome (YAC) -base physical map information of rice from 1994 through 1997 version. |
 | RGP physicalmap2001 |
| Taxonomy Name: Oryza sativa | |
| Description: This database contains the 2001 version of yeast artificial chromosome (YAC) -base physical map information of rice. |
 | RGP kasalath |
| Taxonomy Name: Oryza sativa | |
| Description: This database contains bacterial artifical chromosome (BAC) information of an indica rice variety, Kasalath. You can refer to BAC end sequences and BAC-based physical map information of Kasalath. |
 | RGP chr1 |
| Taxonomy Name: Oryza sativa | |
| Description: This database contains the outline explanation of rice chromosome 1 and links to pertinent data. |